|
 |
- Search
| Healthc Inform Res > Volume 20(2); 2014 > Article |
Abstract
Objectives
The aim of the study was to develop a metadata and ontology-based health information search engine ensuring semantic interoperability to collect and provide health information using different application programs.
Methods
Health information metadata ontology was developed using a distributed semantic Web content publishing model based on vocabularies used to index the contents generated by the information producers as well as those used to search the contents by the users. Vocabulary for health information ontology was mapped to the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), and a list of about 1,500 terms was proposed. The metadata schema used in this study was developed by adding an element describing the target audience to the Dublin Core Metadata Element Set.
As health information provided on the Internet is created by various institutions and individuals with varying degrees of credibility and its target audiences range from the general public to healthcare professionals, the quality and type of information provided also vary widely. In such an environment, it is not easy for ordinary people to find reliable information relevant to their health problems on the Internet. To deal with this issue, health portals, websites that acts as gateways to other sites and aggregate content from multiple resources and present it to the consumers, have emerged. Types of Internet portals include service portals that gather and present various services, community portals that provide virtual meeting places, and information portals that act as data hubs by collecting and presenting data for users.
Health Park (http://www.healthpark.or.kr/) is a health information portal maintained by the Korea Institute for Health and Social Affairs to provide reliable health information to Korean citizens. It presents search results on health information for the promotion and management of Korean citizens' health under cooperative partnership with various health information providers. The search results presented by Health Park include basic search results collected from health-related websites operated by partner institutions and classified by healthcare specialists as well as search results automatically collected and classified by a robot program. Currently, Health Park allows its users to search health and wellness information according to body locations and demographic groups, such as age and sex, as well as in Korean alphabetical order. However, the search capabilities of the Health Park are quite limited compared to those of Medline-Plus, the health information portal operated by the National Library of Medicine in the United States.
In general, when information is published on the Web, content producers create Web pages individually and then link them together. Information portals, such as Health Park, collect and integrate information produced by others and then offer the information to users via link services. Link service to the relevant websites on an information portal is automatically processed by computers, mainly using search engines.
Existing health information portals that collect and present health information for the general public pose various problems for the information users and producers. From the users' perspective, they need to have sufficient knowledge about the institutions that provide content in order to search for relevant content. Meanwhile, from the information producers' perspective, it is difficult and time-consuming to find suitable keywords or metadata to describe the Web pages and documents they produce.
One way to solve these problems is for information portals to use the distributed content generation model that ensures semantic interoperability. By utilizing this model, it is possible to collect and reuse information produced by numerous content producers. Semantic interoperability can be achieved by using a reference terminology model, a standardized clinical terminology system, and knowledge modeling, such as metadata that connects the two [1].
Metadata is structured information that describes, explains, locates, or otherwise makes it easier to retrieve, use, or manage an information resource [2]. Descriptive metadata describes a resource for purposes, such as discovery and identification. It can include elements, such as title, author, abstract, and keywords. Administrative metadata provides information to help manage a resource, such as when and how it was created, file type and other technical information, and who can access it. Descriptive metadata can be created by domain experts who produce information using data standards, such as the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) [3], International Classification of Diseases (ICD) [4], Current Procedural Terminology (CPT) [5], LOINC [6], and HL7 [7]. Administrative metadata can be created by IT specialists who manage automatically collected data using information tools.
The semantic interoperability of a health information search engine can be achieved by using a common ontology to fill values of metadata schema [8]. In the field of health and medical care, ontology has been used in the management of genetic information [9], knowledge base building for medical diagnoses [10], search and classification of disease information [11], and description of nursing processes [12,13,14].
With this background, this study aimed to develop a semantic interoperability ensured health information search engine that efficiently collects health information generated by content producers using a distributed semantic Web content publishing model (Semantic Web, http://www.w3.org) [15] based on a metadata and terminology system and to supply health information using various application programs.
The distributed content creation model was used for the development and reuse of health information in this study. The use of this model was justified as follows. First, this model enables a Web application to reuse health-related content produced by another institution, minimizing redundant work and the cost of content production. Second, this model enables a computer system to automatically maintain semantic links and gather content produced by various content producers, minimizing the maintenance cost of a portal. Since content in a certain subject domain can be created at any time, the system should be able to search and upload the new information on the portal as well as update links to relevant information automatically. Third, the model can provide an intelligent service which enables users to search information they want from their own perspectives and search content using semantic relationships. To achieve this, terminology used in the user interface should comprise terms used by the general public, different from the terminology used by the content providers for indexing the content.
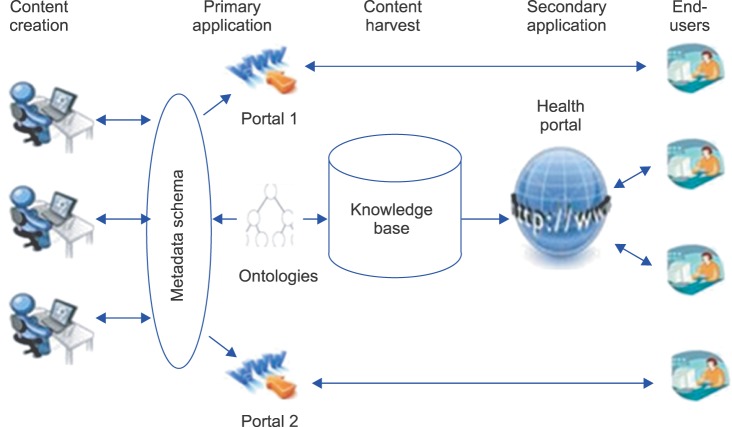
The distributed content creation model is outlined in Figure 1. Content producers shown on the left side of the figure develop and provide information, such as Web pages and documents. This content is indexed using metadata schema and ontology so that others can use it as well. Then, the indexed content is stored in the knowledge base for reuse in the secondary application.
In this study, we aimed to build metadata schema and ontology that can be used for indexing information and apply them to the health information search engine of Health Park. If content collected by Health Park is indexed using metadata and ontology and stored, semantic interoperability of information can be ensured. It would then be possible to share, exchange, and reuse the information.
Web documents to be searched on a health information portal should be described in a format that can be understood by a machine. The metadata schema defines fields (properties) required for presenting information about an individual document. The values of metadata fields are filled with text (e.g., document title) comprehensible to humans, such as structured text strings (e.g., date of publication) or pre-defined ontology concepts (e.g., subject heading). Some of these fields are required fields, and some are recorded more than once. The metadata schema not only provides the format specifications that show the optionality of each field in describing individual documents, but also helps content producers create a suitable content and use it as an interface to test the validity of the content before publishing it.
The metadata schema of the health information search engine developed in this study was based on the Dublin Core Metadata Element Set [16]. Additional elements were identified and added to describe the health information documents in more detail. We specified mandatory elements among these metadata elements.
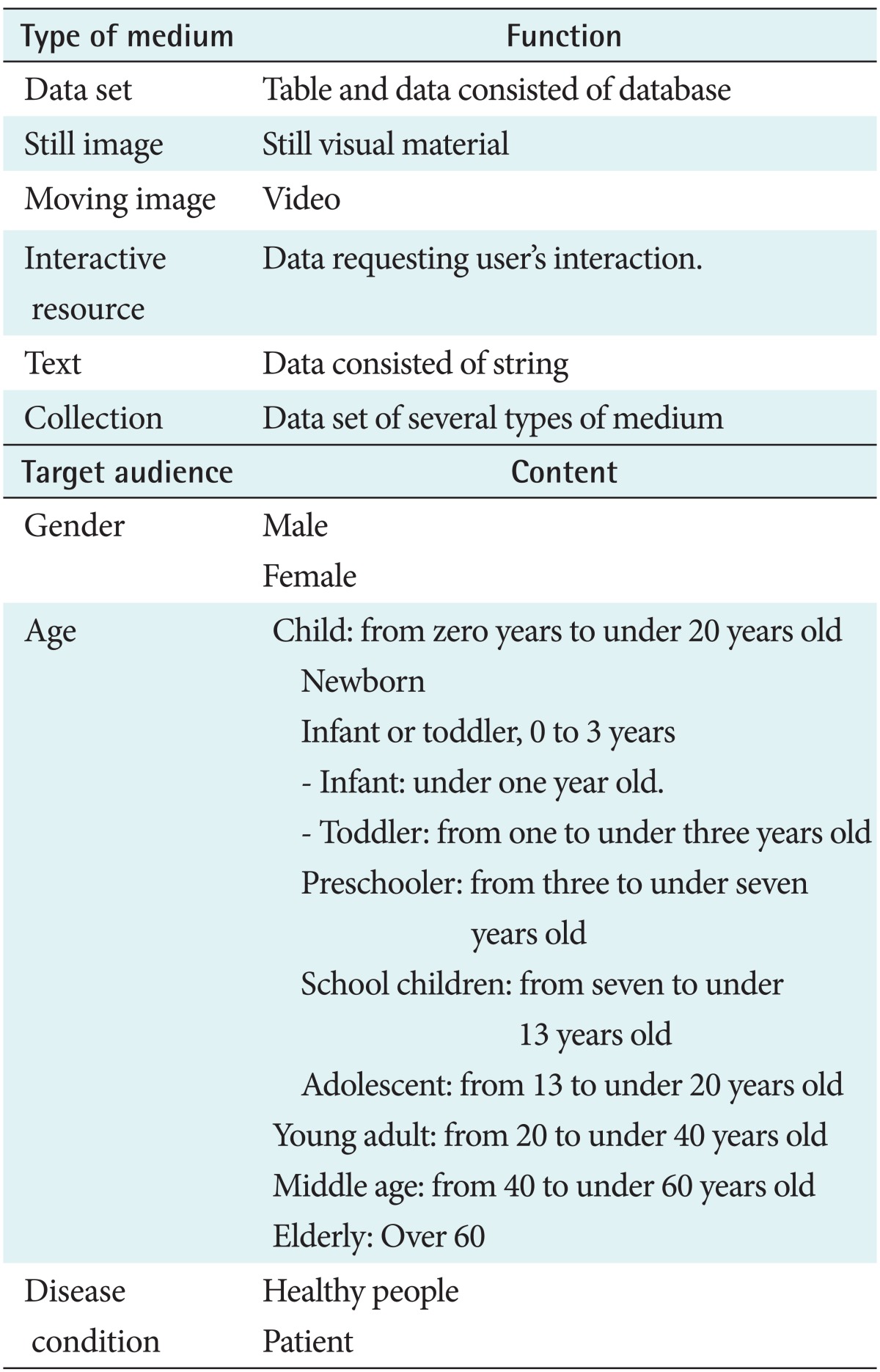
Ontologies to be used for the health information search engine included types of medium of resources, target audience, and subject heading. The ontology for the types of medium used to classify the types of health information was developed using the Type Vocabulary of the Dublin Core Metadata Initiative (DCMI) [17], and the ontology for the target audience was developed with consideration of the concepts used to differentiate health information that is supplied only for specific audience groups. The ontology for the subject heading used to describe the themes of Web content was developed to include the symptoms of health problems, diagnostic methods, treatment methods, and prevention methods.
The vocabulary used to describe the subject heading ontology consists of concepts classifying the content of health information into disease information, health promotion/disease prevention information, and diagnosis/treatment management information. We collected and analyzed vocabulary describing the 'Title/Contents' under the 'Health and Disease Information-Disease/Surgery/Treatment/Diagnostic Tests Information' menu of 'Health iN (http://hi.nhic.or.kr)' portal site operated by the National Health Insurance Cooperation, a partner organization of Health Park. After the morpheme analysis of the collected titles and content, a health information classifier was developed by comparing the analyzed data with the ontology metadata database. This classifier handles analogous terms and synonyms used by the general public.
Commercial search engines, such as Repia External Knowledge Management System (REKMS-3.0) and Repia Search Appliance for website (RSA-3.5) were used as the main solutions for the search engines. Programming language C was used for search engine coding, and JSP was used for the user interface design. Red Hat Enterprise Linux 5.5 (32 bits), 4 GB memory card, SAS 300 GB hard disk drive, MySQL Database ver. 5.0.95, Apache HTTP Server ver. 2.2.3, and Apache Tomcat 6.0.18 were used for the server.
The performance of the search engine was tested through a comparison of the differences between the search results of the existing system and those of the newly developed search system.
The ontology infrastructure of health information search engines consists of a metadata schema and an ontology vocabulary. The metadata schema defines elements to describe Web documents and the types of values the elements can take. The metadata schema can be utilized not only in the extraction and classification of existing health information, but also in the production of health information content by individuals or institutions to guarantee syntactic interoperability of content. The ontology vocabulary system consists of concepts to be used to fill values of metadata schema. If individuals or institutions that develop content use the ontology vocabulary system, the semantic interoperability of content will be ensured.
All elements of the Dublin Core Metadata Element Set (http://dublincore.org/documents/dces/) were used as elements of the metadata schema of the health information search engine in this study. They included the following: identifier, title, description, contributor, language, date, publisher, and creator under the General Metadata category; subject, type, format, and coverage under the Content Classification category; and right, source, and relation under the Relation category. In addition, the target audience was added to the Content Classification category to describe who the main target audiences of the health information documents were (Table 1). We specified identifier and title of General Metadata and subject and type of Content Classification as mandatory elements among these 16 metadata elements.
The ontologies to be used for the health information search engine included the medium types of resources, target audience, and description of the subject heading.
The ontology for the medium type to classify the types of health information was extracted from the DCMI Type Vocabulary [17]. They included data set, still image, moving image, interactive resource, text, and collection of several types of medium (Table 2). The ontology for the target audience included the concepts used for differentiating health information supplied only for special audience groups. They included gender, age, and disease condition (Table 2).
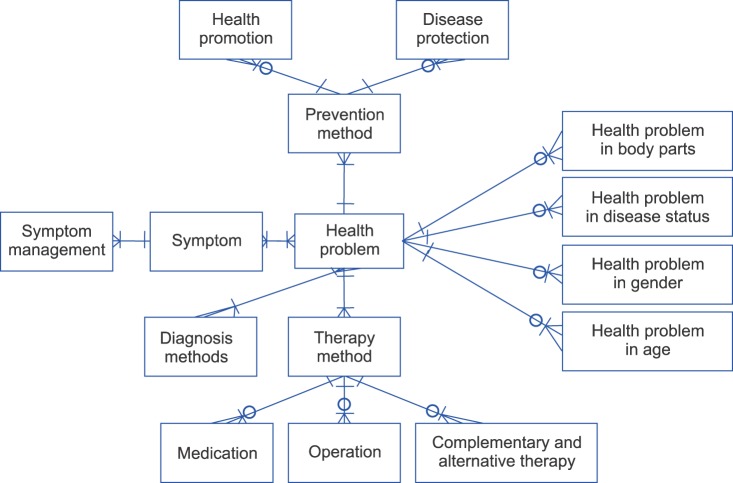
The subject heading ontology was composed of the symptoms of health problems, diagnostic methods, treatment methods, and prevention methods (Figure 2). A given health problem involves one or more symptoms, diagnostic methods, treatment methods, and prevention methods, all of which can be presented according to various aspects, such as relevant body parts, disease status, gender, and age. A specific symptom involves one or more symptom management methods, and a treatment involves various treatment methods, such as medication, operation, and complementary and alternative therapy. A prevention method can also involve different types of health promotion and disease protection methods.
Concerning the cardinality of their relationships, the relationship between a health problem and a symptom is 1:n, the relationship between a health problem and a diagnosis method 1:n, the relationship between a health problem and a therapy method 1:n, the relationship between a health problem and a prevention method 1:n, the relationship between a health problem and body parts 0:n, the relationship between a health problem and a disease status 0:n, the relationship between a health problem and gender 0:n, the relationship between a health problem and age 0:n, the relationship between a symptom and symptom management 1:n, the relationship between a therapy method and a medication 0:n, the relationship between a therapy method and an operation 0:n, the relationship between a therapy method and a complementary/alternative therapy 0:n, the relationship between a preventive method and health promotion 0:n, and the relationship between a preventive method and a disease protection method 0:n.
The vocabulary system used to describe the subject heading consists of concepts classifying the content of health information. They can be grouped into disease information, health promotion/disease prevention information, and diagnosis/treatment management information. Disease information is presented in terms of relevant body parts, disease status, gender, and age groups. Health promotion/disease prevention information incorporates concepts describing actions an individual can take to maintain health and prevent diseases as well as problems faced by family and society as a whole. Diagnosis/treatment information includes concepts describing diagnostic tests, symptom management, and various treatments required when a health problem occurs. The vocabulary system used in this study was mapped to SNOMED CT, and about 1,300 terms were proposed to describe the subject headings of the health information provided on the Internet (Table 3).
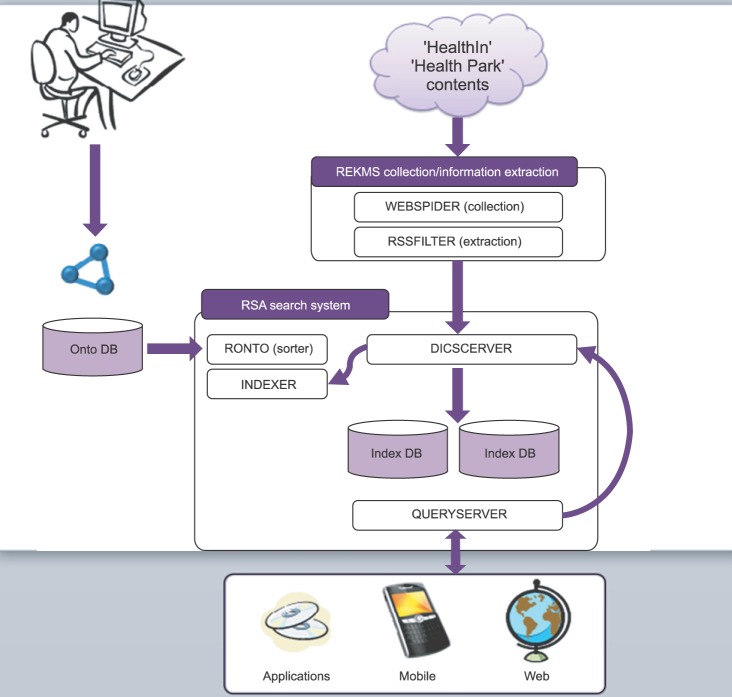
The diagram of the ontology-based health information search system developed in the study is shown in Figure 3. REKMS collects and extracts health information from webpages. The RSA system analyzes a search term entered by a user using mobile and Web applications, matches the analyzed concepts of the search term with health information extracted from the REKMS, and returns ontology-based search results using the Onto DB developed with the metadata subject headings of health information mapped to the SNOMED CT concepts.
A search begins in three different ways. A user can enter a search term into the search Window, select a keyword from the Onto Tree, or enter a search term into the search Window and also select a keyword from the Onto Tree. If the search Window is used, only search results of the search term will appear. If the Onto Tree is used, search results of predefined identifiers will appear. If both the search Window and the Onto Tree are used, search results of both the search term and predefined identifiers will appear. Onto Tree was developed using the metadata title and identifier of the Onto DB.
REKMS collects and extracts health information from the webpages. WebSprider of the REKMS collects source webpages with health information from the Health iN (http://hi.nhic.or.kr) and Health Park. Once the webpage is downloaded, the new source of the webpage in the downloaded webpage is extracted. This process will be repeated until there are no more sources of webpages to collect. The RssFilter of the REKMS extracts the titles and contents of the health information of the webpages after removing unnecessary information and stores them in the database.
When a search term is entered, the QUERYSERVER will send the term to the DICSERVER. The DICSERVER will do a morpheme analysis of the search term and return identifiers of the analyzed concepts. RONTO (Sorter) stores the contents of the Onto DB into memory space to increase search capacity and relieve database loading. The health information of the webpages extracted with the REKMS is searched using the metadata of the RONTO and a priori algorithms. If there is a match, a metadata identifier is assigned to the webpage and stored in the indexing file. One document from the webpage can have more than one metadata identifier.
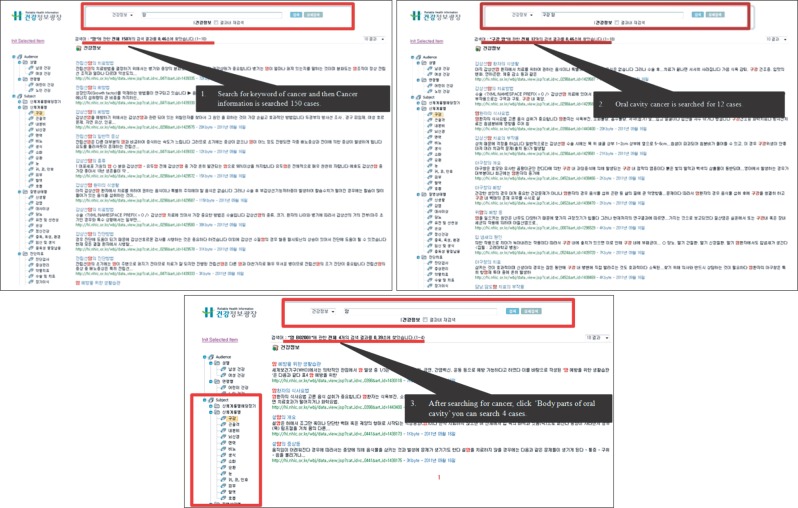
Regarding the performance of the search engine, Figure 4 shows the differences between the search results of the existing system and those of the newly developed 'Ontology-based Health Information Search System.' With the existing system, there were a total of 150 search results when the search was made with the keyword 'cancer,' while there were 12 search results when the search was made with the keywords 'oral + cancer.' On the other hand, 4 search results were found when the keyword 'cancer' was entered and then 'oral' was selected in the Onto Tree.
Content not closely related to oral cancer was presented when the search was made using the keywords 'oral + cancer' with the existing system, while search results for 'tongue
cancer', which is closely related to oral cancer, appeared when the search was made with the newly developed system.
Health information on the Internet is provided in numerous forms by various types of institutions and individuals, including ordinary people and specialists. It is not easy for the ordinary person to find suitable and reliable information that is personally relevant under this environment. Health portals have been introduced to solve these problems. However, these portals often fail to supply the appropriate information that users want.
Existing health information portals that collect and present health information for the general public pose various problems for information users and information producers [18]. Information users experience various types of difficulties when they search for health information on the Internet, such as searching difficulties, connection problems, as well as issues regarding quality and level of specialization. Information producers also find difficulty in producing health information on the Internet, maintaining internal and external links, indexing, managing content quality, and ensuring interoperability.
The greatest obstacle in managing health and medical information is that the processed information is neither structured nor indexed using controlled vocabularies; thus, it is difficult to extract relevant information. To overcome this problem, unstructured text-based content provided on the Internet is processed with a natural language processing application to extract concepts and map them to a standard terminology system, such as SNOMED CT. Then metadata of the concepts is defined and stored in the knowledge base for later use. By utilizing metadata, it is possible to find information resources easily, to structure information based on target audience of the information and themes, and to ensure the interoperability required for information sharing.
With this background, the development of a health information search engine based on metadata ontology is expected to be a new solution that can provide both information producers and users with semantically interoperable health information. Various studies have aimed to develop health information ontology. In particular, the Ontology-based Health Information Inquiry System [19] was built as part of the National Health Promotion Information System to strengthen search capabilities using the health information database system. However, it was limited to a methodology for developing the ontology and failed to actually be imple mented into a service system.
This study aimed to develop a metadata schema and ontology to ensure interoperability in exchanging and communicating health information provided on the Internet, and then developing a metadata schema and ontology-based semantic Web model and portal to create content, collect and provide health information, and implement them in the Health Park search engine.
The metadata schema proposed in the study has 16 elements, with the target audience element added to the 15 metadata elements of the Dublin Core Metadata Element Set which is already widely known, to describe the main target audience of the health information. Among these 16 metadata elements, identifier, title, subject, and type were proposed as mandatory elements.
Also, the health information ontology was divided into health problem, prevention, symptom, diagnosis, and therapy information. These classes can then be divided further. For example, health problem was further classified by body location, disease status, gender, and age. The ontology vocabulary describing subject headings was mapped to SNOMED CT, and a list of about 1,300 terms was suggested to describe health information provided on the Internet.
To develop the ontology-based search engine, first a database was built by collecting concepts describing the 'Title/Contents' of the health and disease information from a health portal operated by the National Health Insurance Cooperation. After the morpheme analysis of the titles and content collected, a health information classifier was developed through a comparison with the ontology metadata database. While the newly developed ontology search engine generated one-third of the search results produced by the existing search engine, the search results were more accurate. The newly developed search engine was able to search a 'tongue cancer' as a type of 'oral cancer'. This was possible because the search keywords were mapped to the ontology with a hierarchical structure.
Therefore, the health information search engine based on meta-ontology that was developed in this study is expected to be a reliable means of providing quality information to information producers and users.
Acknowledgments
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean government (No. 2010-0028631).
References
1. Stroetmann V, Jung B, Rector A, Rodrigues J, Ustun B, Lewalle P, et al. Semantic interoperability deployment and research roadmap (SemanticHealth Project IST 027328 7.1). Brussels: The European Commission; 2007.

2. National Information Standard Organization. Understanding metadata. Bethesda (MD): NISO Press; 2004.

3. International Health Terminology Standard Development Organization. SNOMED CT [Internet]. Copenhagen: International Health Terminology Standard Development Organization; c2014. cited at 2014 Mar 15. Available from: http://www.ihtsdo.org/index.php?id=545

4. World Health Organization. International Classification of Diseases (ICD) [Internet]. Geneva: World Health Organization; c2014. cited at 2014 Mar 15. Available from: http://www.who.int/classifications/icd/en/

5. American Medical Association. Current Procedural Terminology (CPT) [Internet]. Chicago (IL): American Medical Association; c2014. cited at 2014 Mar 15. Available from: http://www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/coding-billing-insurance/cpt.page

6. Regenstrief Institute Inc. LOINC [Internet]. Indianapolis (IN): Regenstrief Institute Inc; c2014. cited at 2014 Mar 15. Available from: http://loinc.org

8. Rector AL. What's in a code? Towards a formal account of the relation of ontologies and coding systems. Stud Health Technol Inform 2007;129(Pt 1):730-734. PMID: 17911813.


9. The Gene Ontology [Internet]. [place unknown]: The Gene Ontology; c2013. cited at 2014 Mar 15. Available from: http://geneontology.org

10. Minchin R, Porto F, Vangenot C, Hartmann S. Symptoms ontology for mapping diagnostic knowledge system Proceedings of the 19th IEEE International Symposium on Computer-Based Medical Systems; 2006 Jun 22-23. Salt Lake City, UT; p. 593-598.

11. Hadzic M, Chang E, Wongthongtham P, Meersman R. Disease ontology based grid middleware for human disease research study Proceedings of the 30th Annual Conference of IEEE Industrial Electronics Society; 2004 Nov 2-6. Busan, Korea; p. 480-486.

12. Kim H, Harris MR, Savova GK, Chute CG. The first step toward data reuse: disambiguating concept representation of the locally developed ICU nursing flowsheets. Comput Inform Nurs 2008;26(5):282-289. PMID: 18769183.



13. Din MA, Abidi SS, Jafarpour B. Ontology based modeling and execution of Nursing Care Plans and Practice Guidelines. Stud Health Technol Inform 2010;160(Pt 2):1104-1108. PMID: 20841855.


14. Hardiker NR. Logical ontology for mediating between nursing intervention terminology systems. Methods Inf Med 2003;42(3):265-270. PMID: 12874660.


15. W3C. Semantic Web [Internet]. [place unknown]: W3C; c2013. cited at 2014 Mar 15. Available from: http://www.w3.org/standards/semanticweb/

16. Dublin Core Metadata Initiative. Dublin Core Metadata Element Set, version 1.1 [Internet]. [place unknown]: Dublin Core Metadata Initiative; c2014. cited at 2014 Mar 15. Available from: http://dublincore.org/documents/dces/

17. Dublin Core Metadata Initiative. DCMI metadata terms [Internet]. [place unknown]: Dublin Core Metadata Initiative; c2014. cited at 2014 Mar 15. Available from: http://dublincore.org/documents/dcmi-type-vocabulary/

-
METRICS

- Related articles in Healthc Inform Res
-
Effects of Health Information Technology on Malpractice Insurance Premiums2015 April;21(2)
Management Issues in Healthcare Information Technology2012 June;18(2)
Implementation of Medical Information Exchange System Based on EHR Standard2010 December;16(4)