I. Introduction
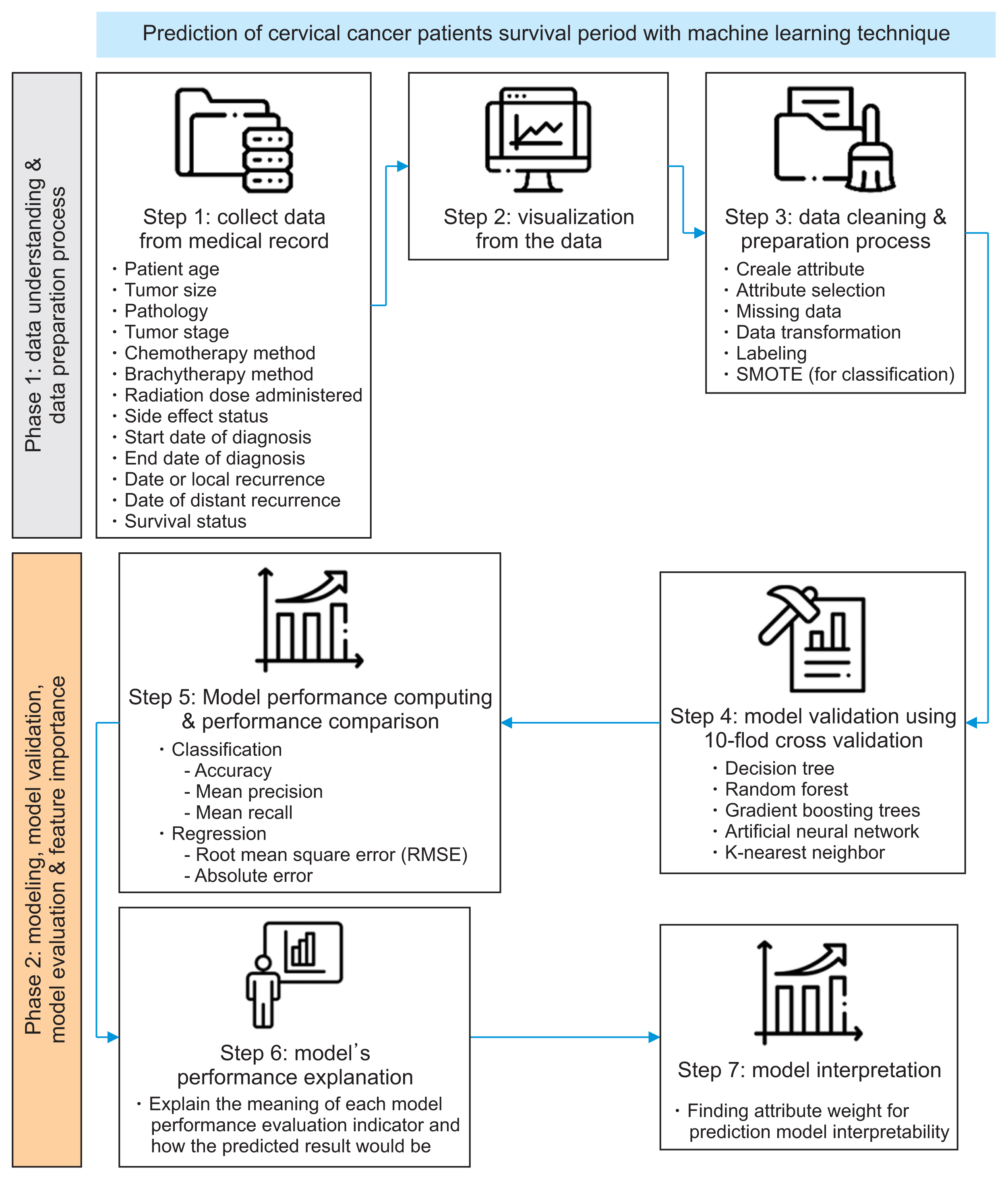
II. Methods
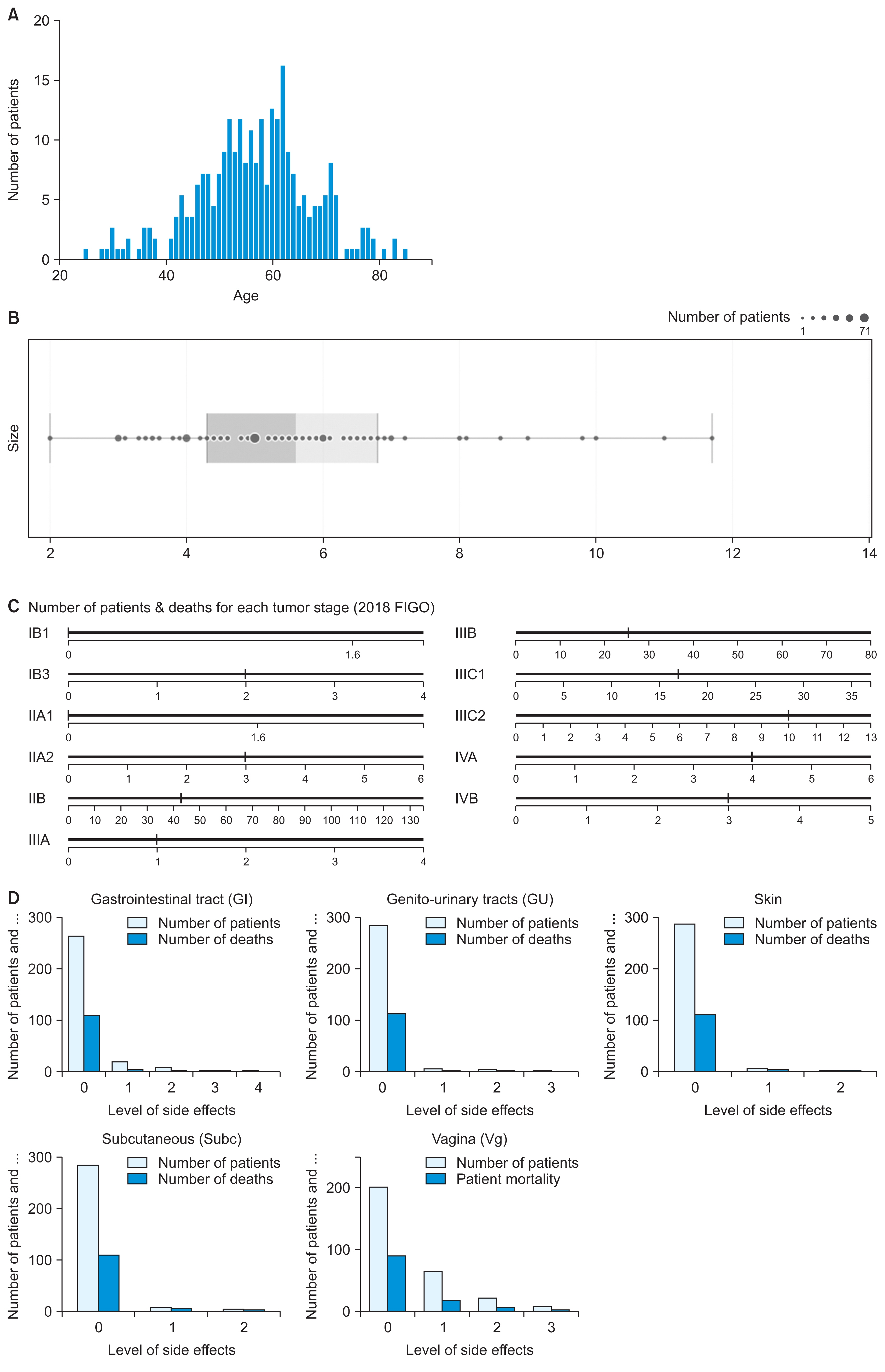
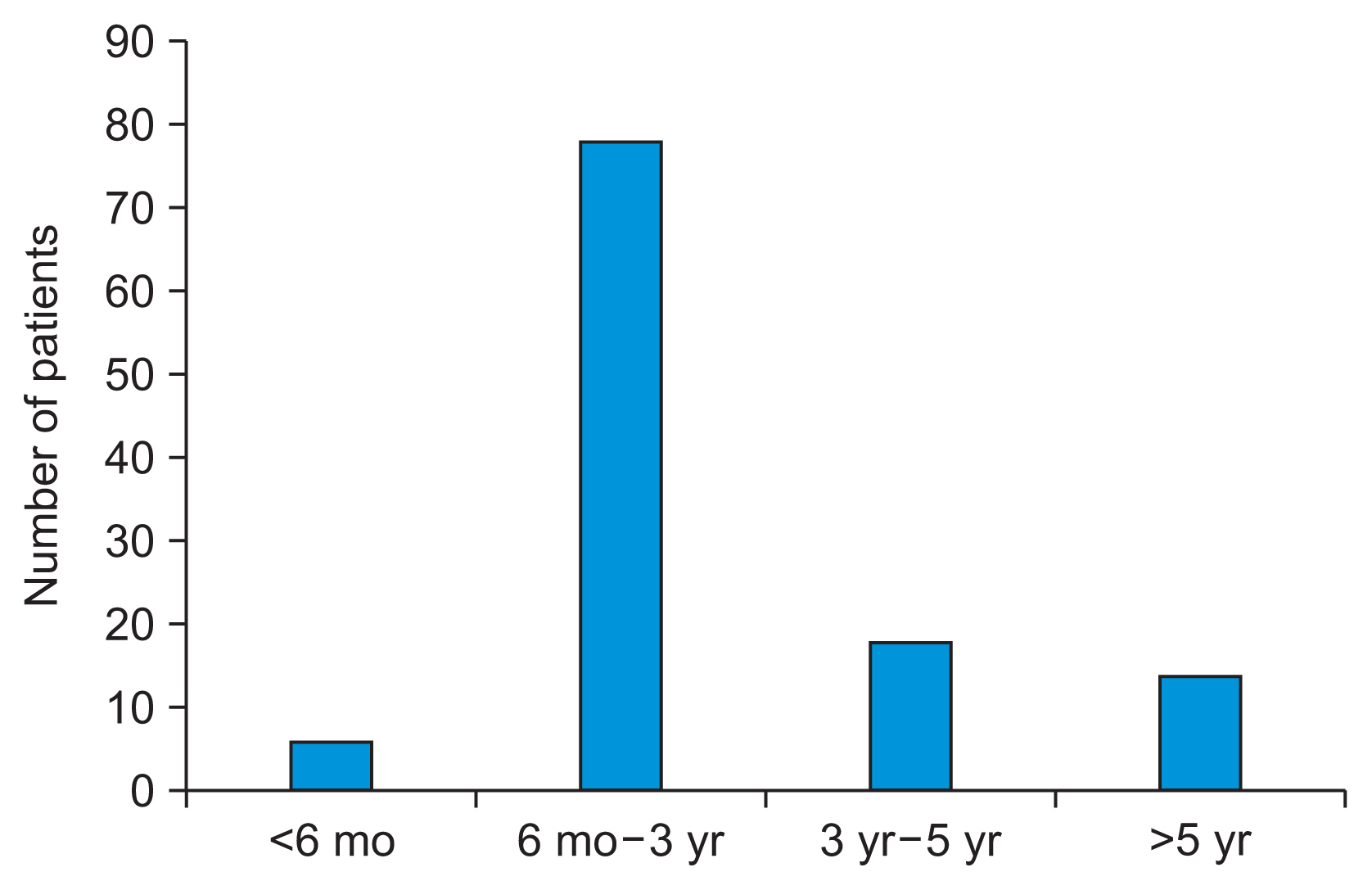
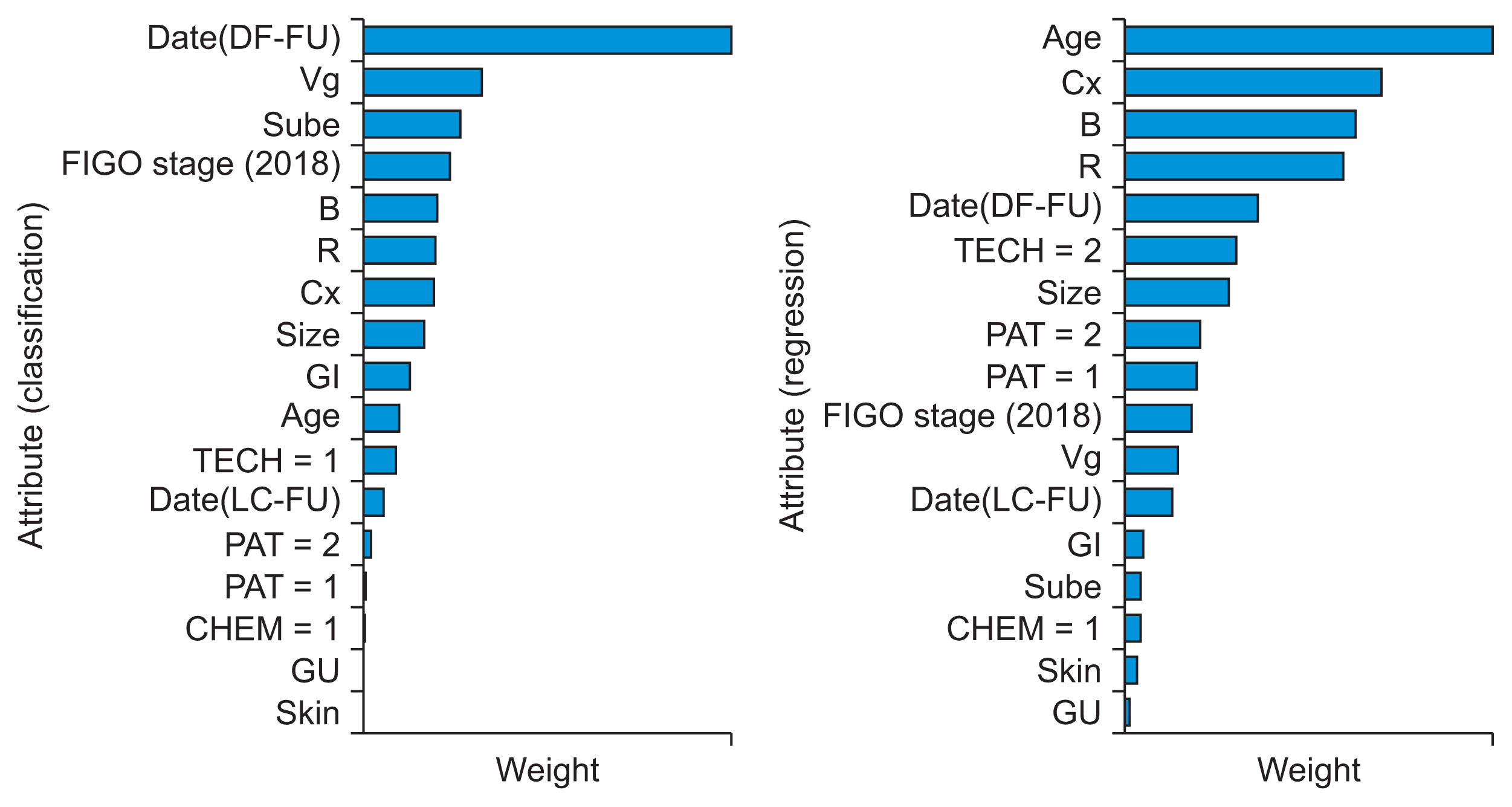
1. Dataset of Cervical Cancer Patients
2. Machine Learning Algorithm
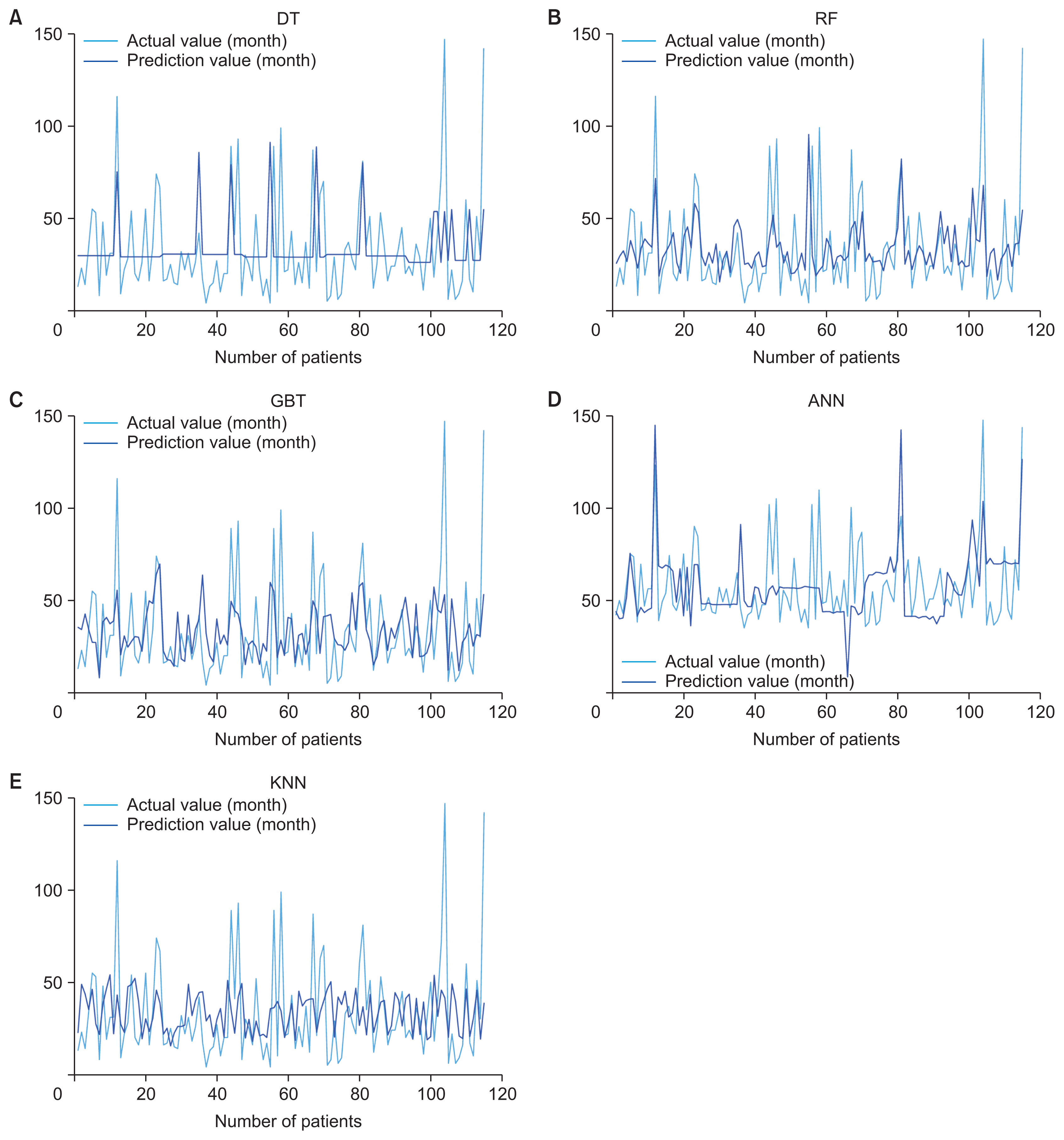
The decision tree (DT) algorithm is widely used in both classification and regression problems. It facilitates hierarchical learning by employing a series of if/else questions that guide the decision-making process. This algorithm comprises three primary components: a node that specifies the attributes of the data; a branch that signifies the test for each attribute value; and a leaf that denotes the outcome for the target class or target values.
The random forest (RF) algorithm is flexible and widely applicable. It operates by constructing an ensemble of decision trees and then assigning the category that garners the majority of votes, or by averaging them, depending on whether the ML modeling is for classification or regression. This approach helps to create a robust model and mitigates the risk of overfitting. In classification modeling, the final prediction is determined by the category that receives the majority of votes from a set of decision trees. Conversely, for regression modeling, predictions are made by averaging the outputs of the decision trees.
-
Gradient boosting tree (GBT) is another widely used boosting algorithm. It utilizes the principle of amalgamating base-level models, which involve multiple simpler algorithms, to develop a better learning algorithm via decision tree construction. This approach is particularly effective for classification and regression modeling. The algorithm iteratively generates trees to correct the errors made by preceding trees, which can be described as follows:
Here, dn (x) is generated by minimizing the error of dn–1 (x) with each sequentially generated tree. -
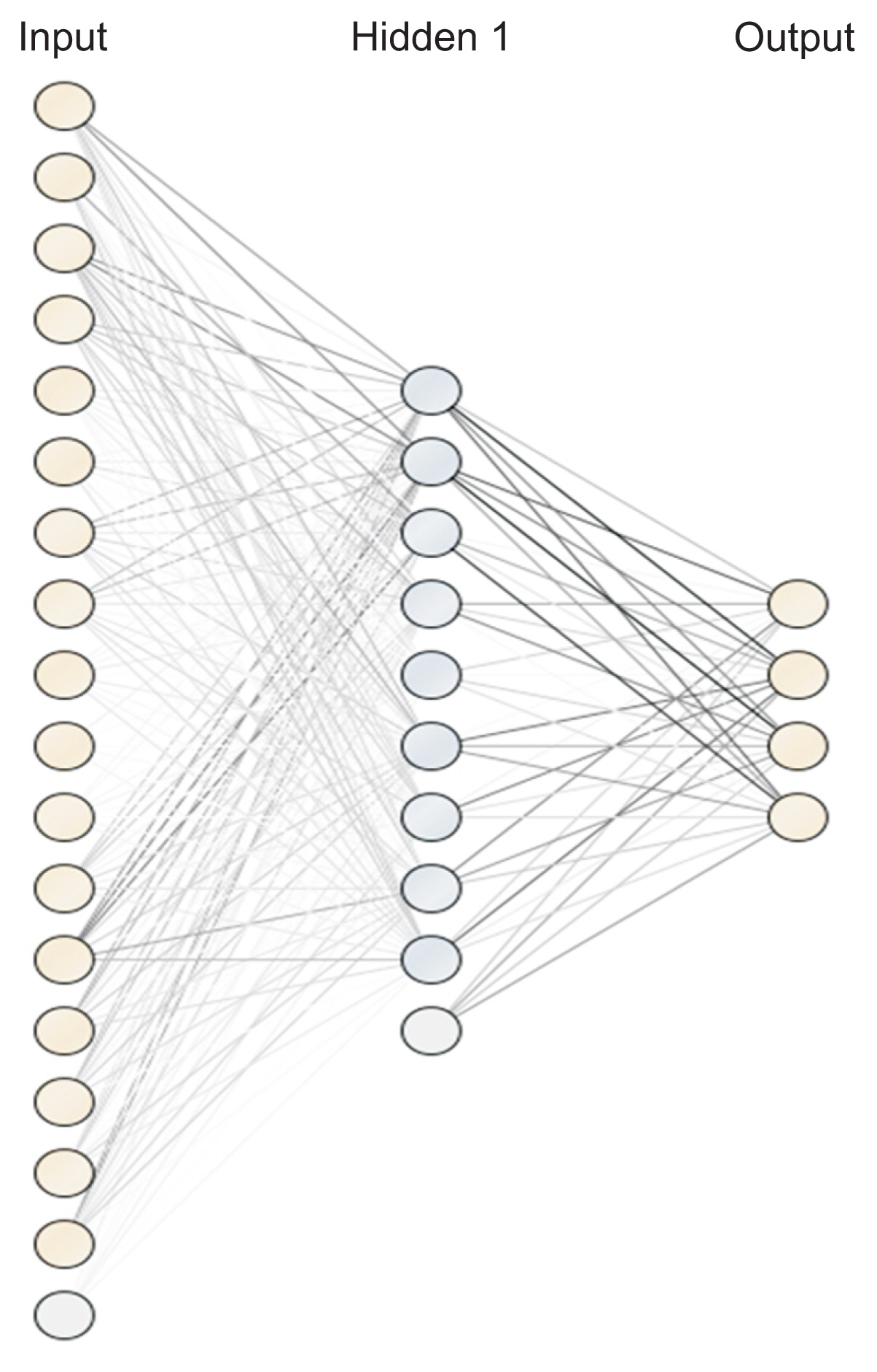
An artificial neural network (ANN) is a method that simulates the function of the nervous system in the form of a network, where nodes are identified as “artificial neurons.” Creating a model of an artificial neuron involves multiplying the inputs by the different weights of each node. Then, calculations are performed using a mathematical function called the activation function to obtain the result. The formula for calculating the weighted sum is shown in following equation:
In this equation, xi represents the input value i and wi represents the weight of input i. The sigmoid function was used as an activation function to determine the output from the node, as shown by the following formula:Then, the backward propagation algorithm was used to improve the weights. The k-nearest neighbor (KNN) method is applicable to both classification and regression problems. It is categorized as a lazy modeling technique because all computations are deferred until after the training phase. To make predictions, KNN calculates the distance between a new data point and those in the training dataset. The decision is influenced by the k nearest data points. A larger k value helps the model minimize the impact of data noise, which can prevent inaccurate results; however, it also increases the risk of overfitting. Euclidean distance is one of the most commonly used metrics for calculating distance in this context.